scientist-run Pathoplexus virus database vows to be transparently run and simple to use
On Aug. 29, 2024, a small team of scientists has launched an open-source database for some of the world’s most dangerous viruses that they say will be run by the very researchers who sequence the pathogens and analyze their genomes.
Called Pathoplexus, the database launched earlier at first will focus on the Sudan and Zaire strains of Ebola virus, as well as Crimean-Congo hemorrhagic fever virus and West Nile virus. Like similar databases, it hopes to help communities derail outbreaks before they grow, and, if that fails, better respond to epidemics and pandemics. But Pathoplexus aims to stand apart in other ways—especially compared with the Global Initiative on Sharing All Influenza Data (GISAID) database, which has become a central repository of sequences for the viruses that cause COVID-19, influenza, mpox, pneumonia, chikungunya, dengue, and Zika.
Pathoplexus will be run by an executive board of sequencing scientists from five continents.Unlike GISAID, Pathoplexus will allow scientists depositing virus sequences to set the terms of whether to make their data fully open or restrict their use. But unlike GISAID, the new database plans to share all unrestricted data it receives with the world’s three main government-funded genome databases (GenBank, EMBL-EBI, and the Database of Japan), as required by many journals publishing analyses of viral sequences. Pathoplexus currently holds fewer than 15,000 sequences, as compared with GISAID’s nearly 18 million.
Tags:
Source: American Association for the Advancement of Science
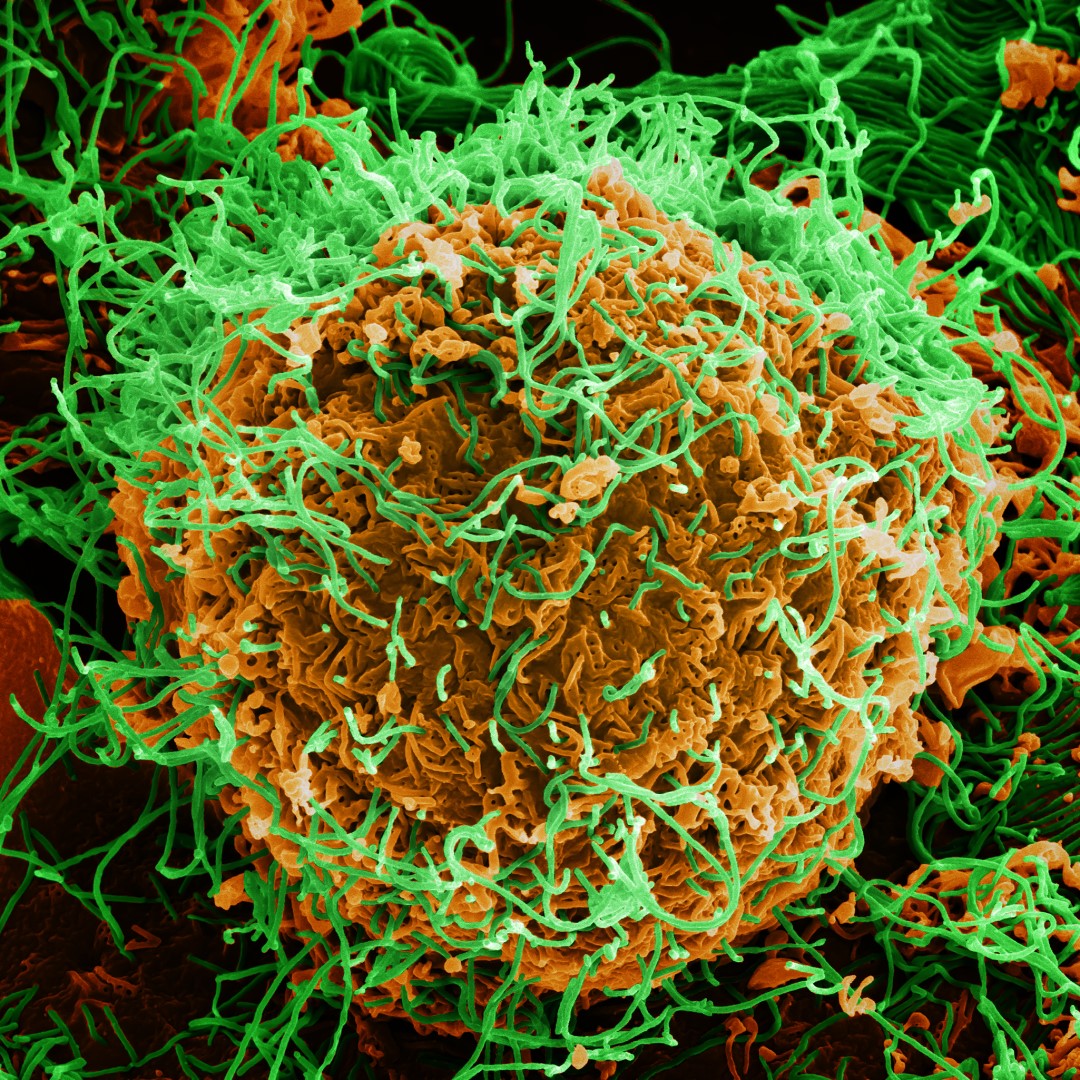
Credit: Photo: Colorized scanning electron micrograph of Ebola virus particles (green) both budding and attached to the surface of infected VERO E6 cells (orange), Fort Detrick, Maryland. Courtesy: National Institute of Allergy and Infectious Diseases.
